(Above images: Created with BioRender.com; Sara Cooper, PhD)
Although many cancers are initially susceptible to chemotherapy, over time they can develop resistance and stop responding to the treatment. For patients with cancers in which chemotherapy is their last or only option, learning their cancer is resistant to treatment is devastating news.
The limited effectiveness of chemotherapy has led to a long effort in the scientific and medical communities to identify alternative treatment strategies. Scientists at the HudsonAlpha Institute for Biotechnology recently contributed to this effort by publishing a paper in BMC Cancer describing the results of a study that identified genes involved in resistance to chemotherapy in pancreatic cancer.
Pancreatic ductal carcinoma and therapeutic resistance
Pancreatic ductal carcinoma (PDAC), also called pancreatic adenocarcinoma, is the most common type of pancreatic cancer, accounting for more than 90 percent of pancreatic cancer diagnoses. The cancer arises in the lining of the ducts in the pancreas, which carry the digestive juices and enzymes to the small intestine to aid in digestion.
Symptoms of PDAC include abdominal and back pain, weight loss, jaundice, and changes to bowel habits. Because of the non-specific symptoms and invasiveness of diagnostic procedures, most patients already have inoperable, locally advanced, or metastatic disease at the time of diagnosis. For these patients, chemotherapy and radiation are generally their sole course of treatment.
PDAC is one of the most lethal diseases, with an average five-year survival of less than 10 percent. The low survival rate can be blamed in part on resistance to therapeutics like chemotherapy. HudsonAlpha Faculty Investigator Sara Cooper, PhD and her lab are focused on discovering genetic bases for chemotherapy resistance in PDAC to help identify new treatment options for these patients.
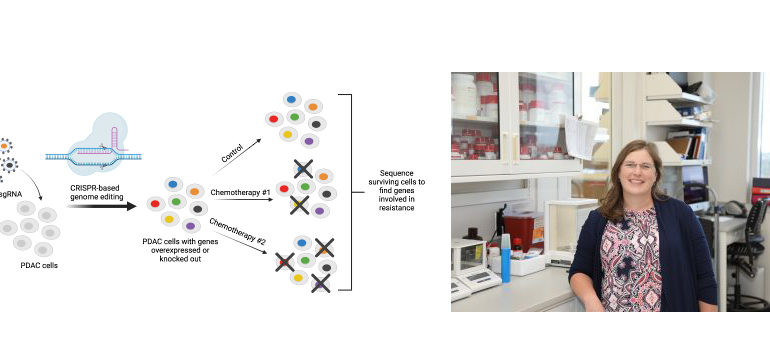
Using CRISPR technology to identify resistance genes
Cooper and her lab used CRISPR gene editing technology to individually activate and knockout each gene in the genome in pancreatic cancer cell lines. They conducted four genome-wide CRISPR activation and knockout screens to identify genes whose loss or gain of expression were able to alter sensitivity to four of the most common chemotherapies used in the treatment of pancreatic cancer (gemcitabine, 5-fluorouracil, irinotecan, and oxaliplatin).
Using these screens, Cooper’s lab identified several gene pathways associated with drug resistance in pancreatic cancer. Activation of chromatin remodeling pathways was one of the most consistent mechanisms of drug resistance identified in the screens and importantly expression of these same genes in patient tumors has been shown to predict survival outcomes.
Chromatin remodeling is the dynamic modification of tightly wound DNA, called chromatin, from its condensed, or closed, state to an open state that allows access to the machinery necessary to turn genes on. Widespread closed chromatin has been previously associated with poor prognosis in pancreatic cancer patients.
The team narrowed in on a gene that codes for an enzyme called histone deacetylase 1 (HDAC1) that is involved in chromatin remodeling. High expression of HDAC1 is also associated with poor patient prognosis in pancreatic cancer. By overexpressing HDAC1 in cell lines, the team discovered that it regulated genes that are part of the epithelial-to-mesenchymal transition, a pathway known to be involved in multi-drug resistance.
“HDAC inhibitors have been used in the treatment of some cancers and are being explored in clinical trials for PDAC however since histone deacetylases are involved in so many functions in the human body, the inhibitors can induce unwanted side effects,” says Cooper. “Now that we have narrowed in on HDAC1 and its involvement in drug resistance, we hope to look at all the genes that are regulated by HDAC1 to see if one or a small subset of them are driving the resistance.”
As part of her graduate work, PhD candidate Carter Wright is continuing to pursue the additional studies aimed at identifying some of the key genes and pathways downstream of HDAC1 that might represent novel therapeutic targets themselves.
Analysis tool to help identify resistance
During the study, the research team sought to assess the clinical relevance of their screening data. Using an algorithm developed in their lab, the group computed scores based on expression levels of resistance genes to predict the sensitivity of PDAC tumors and cell lines to gemcitabine, 5-fluorouracil, irinotecan, and oxaliplatin, four drugs commonly used in PDAC treatment. The algorithm, called PancDS, is freely available for other labs to use in their own pancreatic cancer research.
Although it is solely used for research purposes at this time, the ultimate goal of this type of technology would be to direct personalized therapeutic approaches based upon a tumor’s gene expression profile. For example, if a patient’s tumor expressed high levels of a gene implicated in gemcitabine resistance, then their doctor could prescribe a different treatment regimen.
“The ultimate goal in cancer treatment is to prescribe patients a treatment that will work to eliminate their cancer with minimal side effects,” says Cooper. “By studying gene signatures in tumors, the hope is to be able to predict what a patient’s response to treatment will be, and avoid administering a treatment that won’t work for them and suggest a treatment that is most likely effective.”
About HudsonAlpha: HudsonAlpha Institute for Biotechnology is a nonprofit institute dedicated to developing and applying scientific advances to health, agriculture, learning, and commercialization. Opened in 2008, HudsonAlpha’s vision is to leverage the synergy between discovery, education, medicine, and economic development in genomic sciences to improve the human condition around the globe. The HudsonAlpha biotechnology campus consists of 152 acres nestled within Cummings Research Park, the nation’s second largest research park. The state-of-the-art facilities co-locate nonprofit scientific researchers with entrepreneurs and educators. HudsonAlpha has become a national and international leader in genetics and genomics research and biotech education and fosters more than 45 diverse biotech companies on campus. To learn more about HudsonAlpha, visit hudsonalpha.org.